Today I uploaded the first three lecture chunks (for lack of a better word) of the course I taught this spring on YouTube free for anyone to watch.
The course was offered through the Masters of Science in Biomedical Communications program, the Master's program that I completed two years ago. It covers intermediate and advanced Maya topics, or more specifically, pretty much everything that isn't modelling and texturing.
The recorded lectures are largely Maya demonstrations, tutorials, and explanations that focus on the application of a Maya toolset or technique for biomedical content. Admittedly some of the demos are less biomedical and more "let's use some spheres because it's simpler to demonstrate". They assume some prior knowledge of Maya and a decent understanding of the interface and modelling tools.
Lecture topics (that I will be posting regularly) include Animation, Rigging, nParticles, Optimization, nHair, nCloth, Fluids, Motion Tracking and Compositing with AE, and MEL scripting. I very much hope that you will subscribe to my YouTube channel to view the tutorials as soon as they are posted. There will be 35 videos of varying lengths in the series, so lots of content to come.
Thanks for checking them out! I hope they are informative and useful, and if so, please share them around.
Later,
Stuart
Tuesday, June 30, 2015
Sunday, June 28, 2015
Science Photography EdX Course - Week 1
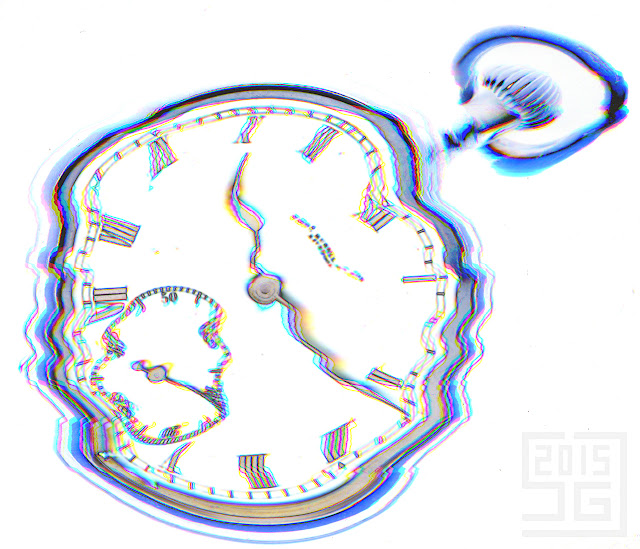
I registered for a Science and Engineering Photography course offered by EdX since I am quite interested in science and nature photography, I've just purchased a macro lens, and I really like the work the instructor has done. Today I completed the first assignment. I will be working with the same object for the duration of the course, so it was important that I choose something interesting and had some significance to me. Fortunately my good wife thought to suggest my great-grandfather's pocket watch, which I inherited after he passed away several years ago at the age of 102. This wasn't his Watch (with a capital W) in the sense of it being the heirloom, since he had several watches and was more of a wrist-watch type of guy, but it still has plenty of significance for me and looking at it now, it could be as old as late 19th century though more likely c. 1930-50. Perhaps I'll be able to discover exactly when it was made while doing this course.
The first assignment was not to use a camera, rather a flatbed scanner. This ended up being a very interesting technique and something I hope to return to in the future. My scanner is capable of scanning at up to 12800 dpi, so ended up with some very large images. A bit overkill, but here is a very scaled down image.
Aside from creating some nice images of the watch face, I stumbled on a pretty surprising effect. Look at the second hand (the inset dial). What is happening here? All the evidence is already in the image, but I'll give a couple of clues. 1) The watch still works 2) The scanner head moves from the top of the image to the bottom. Let me know your analysis in the comments.
And finally, because my artist sister-in-law was over, she noticed the cool moving second hand effect and asked what would happen if I moved the whole watch during the scan. A scan and a bit of photoshop processing later (though not as much as you might expect for this image), I arrived at the below photo.
The first assignment was not to use a camera, rather a flatbed scanner. This ended up being a very interesting technique and something I hope to return to in the future. My scanner is capable of scanning at up to 12800 dpi, so ended up with some very large images. A bit overkill, but here is a very scaled down image.
Aside from creating some nice images of the watch face, I stumbled on a pretty surprising effect. Look at the second hand (the inset dial). What is happening here? All the evidence is already in the image, but I'll give a couple of clues. 1) The watch still works 2) The scanner head moves from the top of the image to the bottom. Let me know your analysis in the comments.
And finally, because my artist sister-in-law was over, she noticed the cool moving second hand effect and asked what would happen if I moved the whole watch during the scan. A scan and a bit of photoshop processing later (though not as much as you might expect for this image), I arrived at the below photo.
I just love the swirly chromatic strands on the hour and minute hands.
This course, while very exciting and about which I will be posting frequently, is not the special announcement I was going to make, so stay tuned again for the actual announcement.
Later,
Stuart
Saturday, June 27, 2015
Molecular Visualization Principles #1-3
As a part of my research work, we are developing some principles, guidelines, or considerations for visualizing molecules in biological systems. It's hard to know exactly how to phrase and frame these, because they are not hard and fast rules; however, they may be helpful in creating more accurate depictions of biomolecules.
Each principle is presented as a short pair of animations. The Treatment A's do not adhere to the principle whereas the Treatment B's do. So far, I have created examples to demonstrate three of the principles, with more coming.
Principle 01 is "Leave some behind". It conveys the idea that typically not all molecules are consumed in a process. For example, there may be more subunits present in the environment than ultimately bind together in a biological complex. This example shows a virus capsid assembling. Note that many of the actual mechanics of capsid assembly (i.e. ideas that may constitute other principles) are ignored or simplified for the sake of clarity. One might declare some irony or hypocrisy here, however it is important that we build the framework for the principles individually before considering how they might be combined in real-world visualization cases.
Principle 02 is "Use Random Walks". It conveys the idea that molecules move randomly due to collisions in a crowded environment. For example, an Arp2/3 complex moves erratically before binding to an actin filament.
Principle 03 is "Add Non-binding Collisions". It conveys the idea that molecules might encounter each other with unproductive orientations and conformations multiple times before a successful binding event occurs. For example, Sos and Ras might collide a number of times before their orientations result in tight binding.
I'll be announcing an exciting new set of content very soon, so check back in a couple of days for that.
Later,
Stuart
Each principle is presented as a short pair of animations. The Treatment A's do not adhere to the principle whereas the Treatment B's do. So far, I have created examples to demonstrate three of the principles, with more coming.
Principle 01 is "Leave some behind". It conveys the idea that typically not all molecules are consumed in a process. For example, there may be more subunits present in the environment than ultimately bind together in a biological complex. This example shows a virus capsid assembling. Note that many of the actual mechanics of capsid assembly (i.e. ideas that may constitute other principles) are ignored or simplified for the sake of clarity. One might declare some irony or hypocrisy here, however it is important that we build the framework for the principles individually before considering how they might be combined in real-world visualization cases.
Principle 02 is "Use Random Walks". It conveys the idea that molecules move randomly due to collisions in a crowded environment. For example, an Arp2/3 complex moves erratically before binding to an actin filament.
Principle 03 is "Add Non-binding Collisions". It conveys the idea that molecules might encounter each other with unproductive orientations and conformations multiple times before a successful binding event occurs. For example, Sos and Ras might collide a number of times before their orientations result in tight binding.
I'll be announcing an exciting new set of content very soon, so check back in a couple of days for that.
Later,
Stuart
Subscribe to:
Posts (Atom)